Project summary
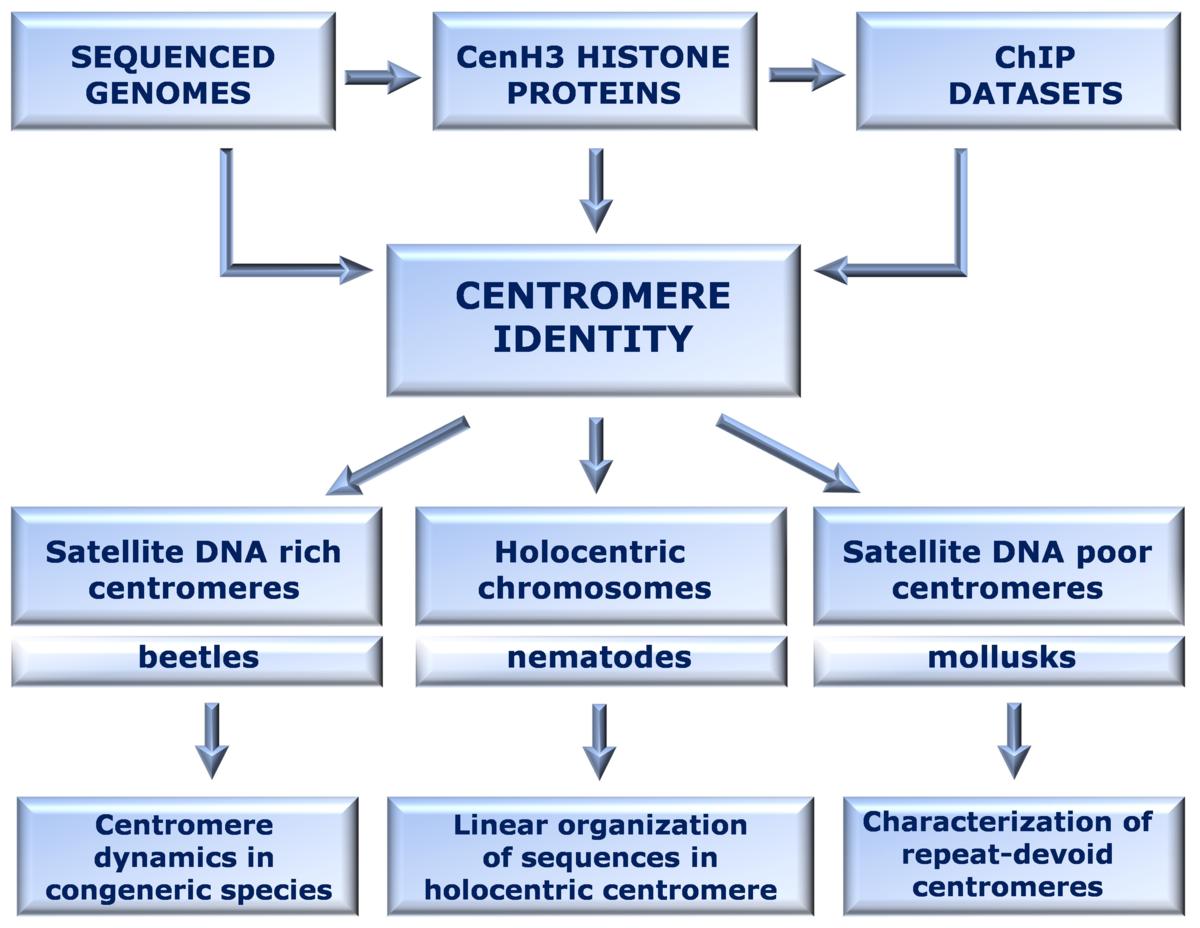
Centromere is a domain essential for chromosomal segregation in cell divisions. Understanding its structure and function is of the highest importance in biology, and also crucial from biomedical point of view. Centromere identity involves both genomic and epigenetic mechanisms, favoring rapid co-evolution and species specificity of CenH3 and associated DNA sequences without impairing the function. Despite diversity in DNA sequences and sequence types, the most common centromeric DNA components in many animal and plant species are repetitive sequences such as satellite DNAs and transposable elements.
Complexity and variations make perception of centromere genomics still fragmental and worked out only in a few most common plant and animal model systems. The main goal of this research is to identify CenH3 and associated DNA sequences in order to explore genomics of differently organized centromeres: satellite DNA-rich, satellite DNA-poor and dispersed on holocentric chromosomes.
Model organisms
The research is focused on the three invertebrate model organisms:
1) the beetle Tribolium castaneum, having centromeres deeply embedded into the satDNA arrays of pericentromeric heterochromatin
2) the root-knot nematode Meloidogyne incognita, possessing holocentric chromosomes
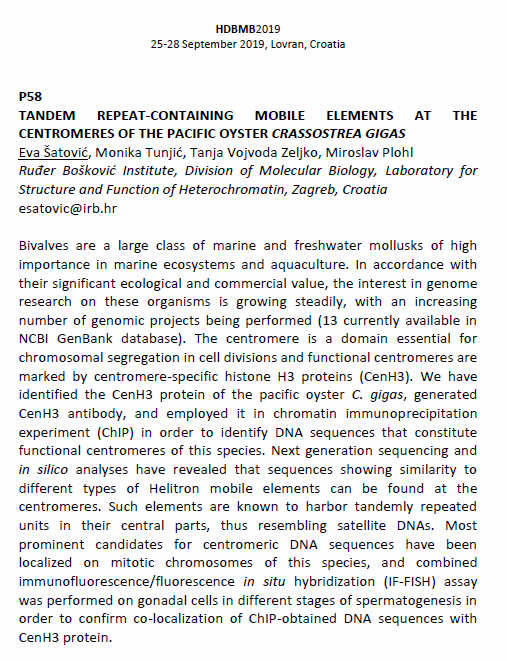
3) the mollusk Crassostrea gigas, lacking abundant and uniformly distributed satDNAs (and/or heterochromatin) in centromeric regions.
Methods
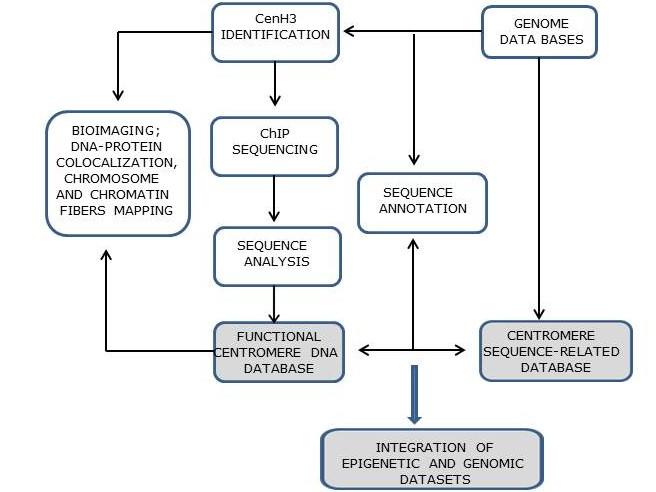
The research strategy combines CenH3 identification, chromatin immunoprecipitation (ChIP)-based assays using anti-CenH3 antibodies, fluorescence in situ hybridization (FISH), immunofluorescence in situ localization (IF) and the bioinformatics pipeline.
Equipment obtained during project implementation
Cytocentrifuge Shandon Cytospin4
Selected publications
Šatović E, Luchetti A, Pasantes J J, García-Souto D, Cedilak A, Mantovani B, Plohl M (2019) Terminal-Repeat Retrotransposons in Miniature ( TRIMs ) in Bivalves. Scientific Reports 9 (19962). https://doi.org/10.1038/s41598-019-56502-y
García-Souto D, Mravinac B, Šatović E, Plohl M, Moran P, Pasantes JJ (2017) Methylation profile of a satellite DNA constituting the intercalary G+C-rich heterochromatin of the cut trough shell Spisula subtruncata (Bivalvia, Mactridae). Scientific Reports 6930; 1-12 DOI:10.1038/s41598-017-07231-7
Šatović E, Vojvoda Zeljko T, Luchetti A, Mantovani B, Plohl M (2016) Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genomics 17:997 DOI 10.1186/s12864-016-3347-1
Luchetti A, Šatović E, Mantovani B, Plohl M (2016) RUDI, a short interspersed element of the V-SINE superfamily widespread in molluscan genomes. Mol Genet Genomics 291:1419–1429. DOI 10.1007/s00438-016-1194-z
Pavlek M, Gelfand Y, Plohl M, Meštrović N (2015) Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Research. 22:387–401. DOI: 10.1093/dnares/dsv021
Meštrović N, Mravinac B, Pavlek M, Vojvoda-Zeljko T, Šatović E, Plohl M (2015) Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 23:583-596. DOI: 10.1007/s10577-015-9483-7.
Plohl M, Meštrović N, Mravinac B (2014) Centromere identity from the DNA point of view. Chromosoma, 123:313-325. DOI: 10.1007/s00412-014-0462-0
Šatović E, Plohl M (2013) Tandem repeat-containing MITEs in the clam Donax trunculus. Genome Biol Evol 5 (12): 2549-2559. DOI: 10.1093/gbe/evt202.
Meštrović N, Pavlek M, Car A, Castagnone-Sereno P, Abad P, Plohl M (2013) Conserved DNA motifs, including the CENP B box-like, are possible promoters of satellite DNA array rearrangements in nematodes. PLoS ONE 8(6): e67328.
Mravinac B, Meštrović N, Čavrak V, Plohl M (2011) TCAGG, an alternative telomeric sequence in insects. Chromosoma, 120:367-376.
Mravinac B, Plohl M. (2010) Parallelism in evolution of highly repetitive DNAs in sibling species. Mol Biol Evol, 27:1857-1867. DOI: 10.1093/molbev/msq068
Plohl M, Petrović V, Luchetti A, Ricci A, Šatović E, Passamonti M, Mantovani B. (2010) Long-term conservation vs. high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 104:543-551. DOI:10.1038/hdy.2009.141
Meštrović, N., Plohl, M., Castagnone-Sereno, P. (2009) Relevance of satellite DNA genomic distribution in phylogenetic analysis: a case study with root-knot nematodes of the genusMeloidogyne. Mol. Phyl. Evol. 50, 204-208. DOI: 10.1016/j.ympev.2008.10.013
Plohl, M., Luchetti, A., Meštrović, N., Mantovani, B. (2008) Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric heterochromatin. Gene 409, 72-82
Meštrović N, Castagnone-Sereno P, Plohl M. (2006) Interplay of selective pressure and stochastic events directs evolution of the MEL 172 satellite DNA library in root-knot nematodes. Mol Biol Evol 23:2316-2325.
Meštrović, N., Plohl, M., Mravinac, B. and Ugarković, Đ. (1998) Evolution of satellite DNAs from the genus Palorus- experimental evidence for the "library" hypothesis. Mol. Biol. Evol. 15, 1062-1068
Dissemination of Results
Congress of the Croatian Society of Biochemistry and Molecular Biology, HDBMB2019, "Crossroads in Life Science", Lovran, Croatia, 25.-28.9. 2019.
Invited oral presentation abstract
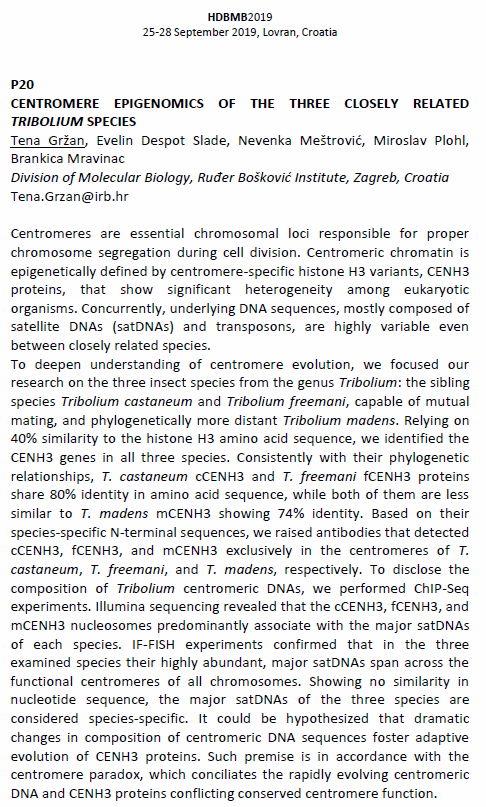
Poster presentation abstract
Poster presentation abstract
13th CROATIAN BIOLOGICAL CONGRESS WITH INTERNATIONAL PARTICIPATION, POREČ, CROATIA, 19.-23.9.2018.
Invited oral presentation abstract
SATELLITE DNA SEQUENCES IN CENTROMERIC CHROMOSOMAL REGIONS
M. PlohlRuđer Bošković Institute, Bijenička c. 54, HR-10000 Zagreb, Hrvatska (plohl@irb.hr)
Satellite DNAs are non-coding DNA sequences that build long arrays of tandem repeats usually located in heterochromatic compartments of eukaryotic chromosomes. They represent preferred genome component in pericentromeric regions, spanning also the functional centromere. Each genome harbors many satellite DNAs which differ in sequence, copy number and chromosomal localization. According to the library model, species-specific differences in satellite DNA composition evolve rapidly as a consequence of copy number alterations within a library of satellite DNAs, rather than as a consequence of changes in nucleotide sequences. This feature of satellite DNA sequence dynamics might be advantageous in maintaining DNA-protein interactions in centromeres. Since there is no particular centromeric DNA, the most prominent species-specific marker of centromere function is histone H3 protein variant CenH3, N-terminal part of which is associated with the DNA sequence. As a consequence, CenH3 and centromeric DNA sequences must remain in accord, and they therefore drive each other’s evolution. Accumulation of differences in DNA/protein components among groups of individuals can cause incompatibilities in chromosome pairing in hybrids, trigger reproductive isolation and ultimately speciation. From the genomic point of view, (peri)centromeres are the least known regions, underrepresented in outputs of every genome project because of difficulties in sequencing and assembly of long arrays of repetitive DNAs. Advances in NGS sequencing and in analysis of repetitive sequences opened possibility to study the satellitome, the whole-genome complement of satellite DNAs, and together with the immunoprecipitation method to focus onto detailed composition of DNA sequences in the centromere.
Poster presentation abstract
LOW-COPY SATELLITE DNAs OF THE FLUOR BEETLE TRIBOLIUM CASTANEUM REVEALED BY NEXT-GENERATION SEQUENCING
T. Gržan1, M. Dombi1, M. Plohl1, B. Mravinac1
1 Division of Molecular Biology, Ruđer Bošković Institute, Bijenička street 54, 10000 Zagreb, Croatia
Satellite DNAs (satDNAs) are tandemly repeated sequences prevalent in (peri)centromeric chromosomal regions that represent one of the most enigmatic parts of eukaryotic genomes. The red flour beetle Tribolium castaneum is an excellent model organism used in molecular studies for decades. Ten years ago its genome was sequenced by a shotgun approach; however, 20% of the genome is omitted from the current assembly due to the repetitive set-up based primarily on satDNAs. In course of the study of centromeric DNA sequences in T. castaneum, we re-sequenced its genome by Illumina Next-Generation Sequencing (NGS). The satellitome, representing the complete set of satDNAs in the genome, was analyzed by a computational pipeline TAREAN, which enables detection of putative satellite sequences from unassembled short reads. We identified three novel T. castaneum satDNAs that have not been detected in the previous studies. In spite of low contribution to the genome (<0.1%), we successfully proved tandem organisation of these repeats by Southern blot method. Fluorescence in situ hybridization also revealed their localization on different chromosomal subsets. By combining bioinformatics tools and experimental procedures we affirmed the applicability of NGS unassembled reads in detection and characterization of low-copy satDNAs, which might be relevant to understanding genome structure and function.
4th CONGRESS OF CROATIAN GENETICISTS WITH INTERNATIONAL PARTICIPATION, Krk, Croatia, 26.-29.9.2018
Oral presentation abstract
ROOT-KNOT NEMATODES AS MODEL FOR CENTROMERE AND SATELLITOME RESEARCH
Despot Slade Evelin1, Martina Pavlek1, Mravinac Brankica1, Castagnone-Sereno Phillipe2, Plohl Miroslav1, Meštrović Nevenka1
1 Division of Molecular Biology, Ruđer Bošković Institute, Bijenička street 54, 10000 Zagreb, Croatia
2 INRA/UNSA/CNRS, Institut Sophia Agrobiotech, 400 Route des Chappes, BP167, 06903 Sophia Antipolis, France
Root-knot nematodes (genus Meloidogyne) are established as important model organisms of plant parasites. Their specific characteristics such as holocentric chromosomes, polyploidy and parthenogenesis make them an ideal system to study the genome evolution, speciation processes as well as centromere structure and function. In this regard, our analysis of centromere specific histone H3 (CENH3) reveal three divergent CENH3 variants per genome which are completely conserved in related species. Transcriptome data show developmental stage-specific expression of CENH3 variants. Immunostaining of chromosomes with antibodies specific for CENH3 variants reveals holocentric distribution pattern. Given that the centromere tend to be established upon satellite DNA arrays even in some holocentric species we further focus on the characterization of satellite DNAs in related Meloidogyne genomes. Using next-generation sequencing combined with novel bioinformatics tools we detected remarkable collection of different satDNAs (satellitome) in each genome. Interestingly, comparative analyses of Meloidogyne satelitomes show set of satDNAs common for all analysed species, set shared by three related species and set of satDNAs conserved in sister species. All results will be discussed in terms of coexistence of different CENH3 variants in the genome, their role in holocentric centromere together with trends in satellitome evolution and compare with the recently established hypothesis according to which the origin of Meloidogyne species is result of multiple hybridisation events.
Poster presentation abstract
COMPARATIVE SATELLITOME ANALYSIS REVEALS THE POLYPLOID HYBRID ORIGIN IN HOLOCENTRIC NEMATODES
Despot Slade Evelin1, Širca Saša2, Plohl Miroslav1, Meštrović Nevenka1
1 Division of Molecular Biology, Ruđer Bošković Institute, Bijenička street 54, 10000 Zagreb, Croatia
2 Agricultural institute of Slovenia, Ljubljana, Slovenija
Satellite DNAs (satDNAs) are tandemly repeated non-coding DNA sequences, and the most common constituents of every functional centromere. Holocentric chromosomes and multiple genome copies establish nematodes of the genus Meloidogyne as an attractive model for studying centromere-associated DNA sequences. In order to identify a pool of satDNAs that may be involved in genomics of the holocentromere, we have analyzed the satellitome (whole-genome complement of satDNAs) in four different publicly available Meloidogyne genomes using RepeatExplorer computational tool. Our results of genome-wide analyses revealed the presence of a remarkable collection of 38-80 different satDNAs in each examined genome. In addition, comparative analyses of satelitomes show three main groups of satDNAs; common for all four studied species, and shared by subsets of three or only two species. This distribution of satDNAs supports a recent hypothesis based on comparative analyses of coding regions, that speciation processes in Meloidogyne may be a result of additive interspecific hybridizations. Our preliminary analysis of satDNA distribution on chromosomes of studied species also speaks in favor of such unusual genome evolution in Meloidogyne.
22nd INTERNATIONAL CHROMOSOME CONFERENCE, 2.-5.9.2018., Prague, Czech Republic
Poster presentation abstract
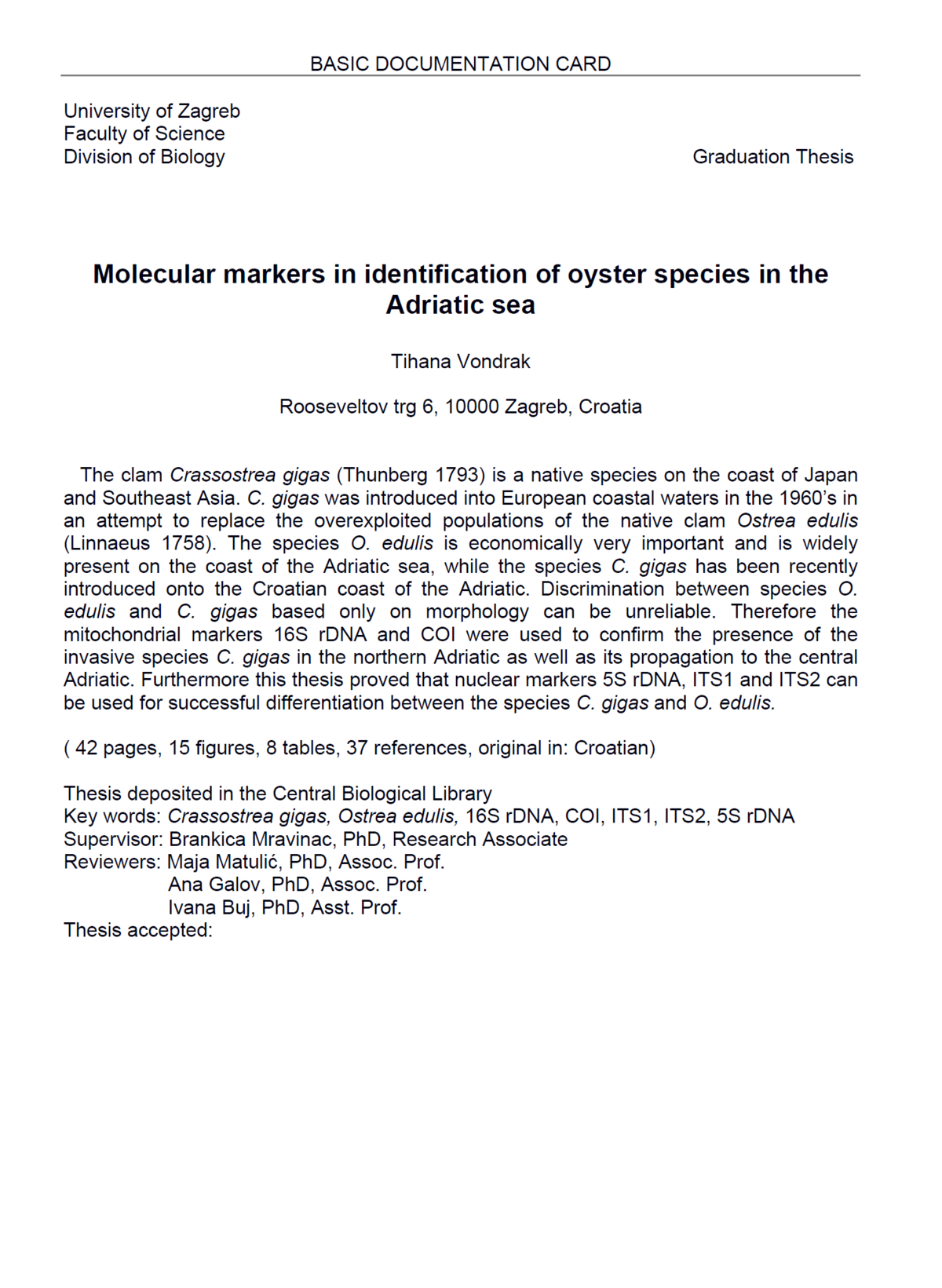
Graduation Theses Obtained During Project Implementation
Project publications
Tunjić Cvitanić, M.; Vojvoda Zeljko, T.; Pasantes, J. J.; García-Souto, D.; Gržan, T.; Despot-Slade, E.; Plohl, M.; Šatović, E. Sequence Composition Underlying Centromeric and Heterochromatic Genome Compartments of the Pacific Oyster Crassostrea Gigas. Genes (Basel). 2020, 11 (6), 695. https://doi.org/https://doi.org/10.3390/genes11060695
Gržan, Tena; Despot-Slade, Evelin; Meštrović, Nevenka; Plohl, Miroslav; Mravinac, Brankica. CenH3 distribution reveals extended centromeres in the model beetle Tribolium castaneum // Plos genetics, 16 (2020), 10; e1009115, 28 https://doi.org/10.1371/journal.pgen.1009115
Vojvoda Zeljko, Tanja; Pavlek, Martina; Meštrović, Nevenka; Plohl, Miroslav. Satellite DNA‑like repeats are dispersed throughout the genome of the Pacific oyster Crassostrea gigas carried by Helentron non‑autonomous mobile elements // Scientific reports, 10 (2020), 15107, 12 https://www.nature.com/articles/s41598-020-71886-y
Positions available
To find out more about opportunities in LSFH contact: plohl@irb.hr
Funding
The project is funded by Croatian Science foundation http://www.hrzz.hr/default.aspx?id=47