Tandemly repeated DNA sequences, known as satellite DNAs (satDNAs), constitute significant portions of eukaryotic genomes, being usually located in genetically silent heterochromatin.
SatDNA biological relevance has been disregarded for decades mostly due to their non-coding character, but it is coming to light that these sequences play important roles in centromere structure, chromatin remodelling, speciation processes, as well as in tumorigenesis. Repetitiveness of satDNAs, that impedes contiguous sequence assemblies, has kept them underrepresented in genome project outputs.
Recently, however, high-throughput sequencing accompanied by specialized computational tools has revolutionized satDNA studies enabling discovery of a whole set of satDNAs in a genome, named a satellitome.
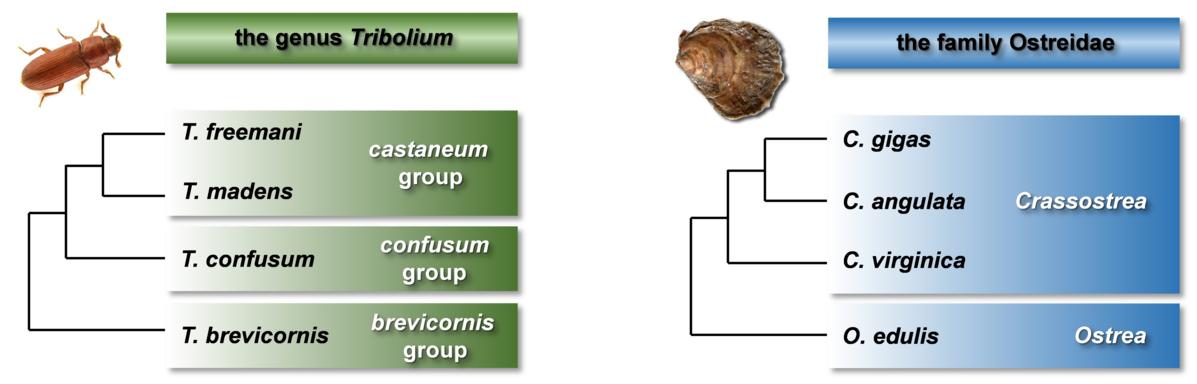
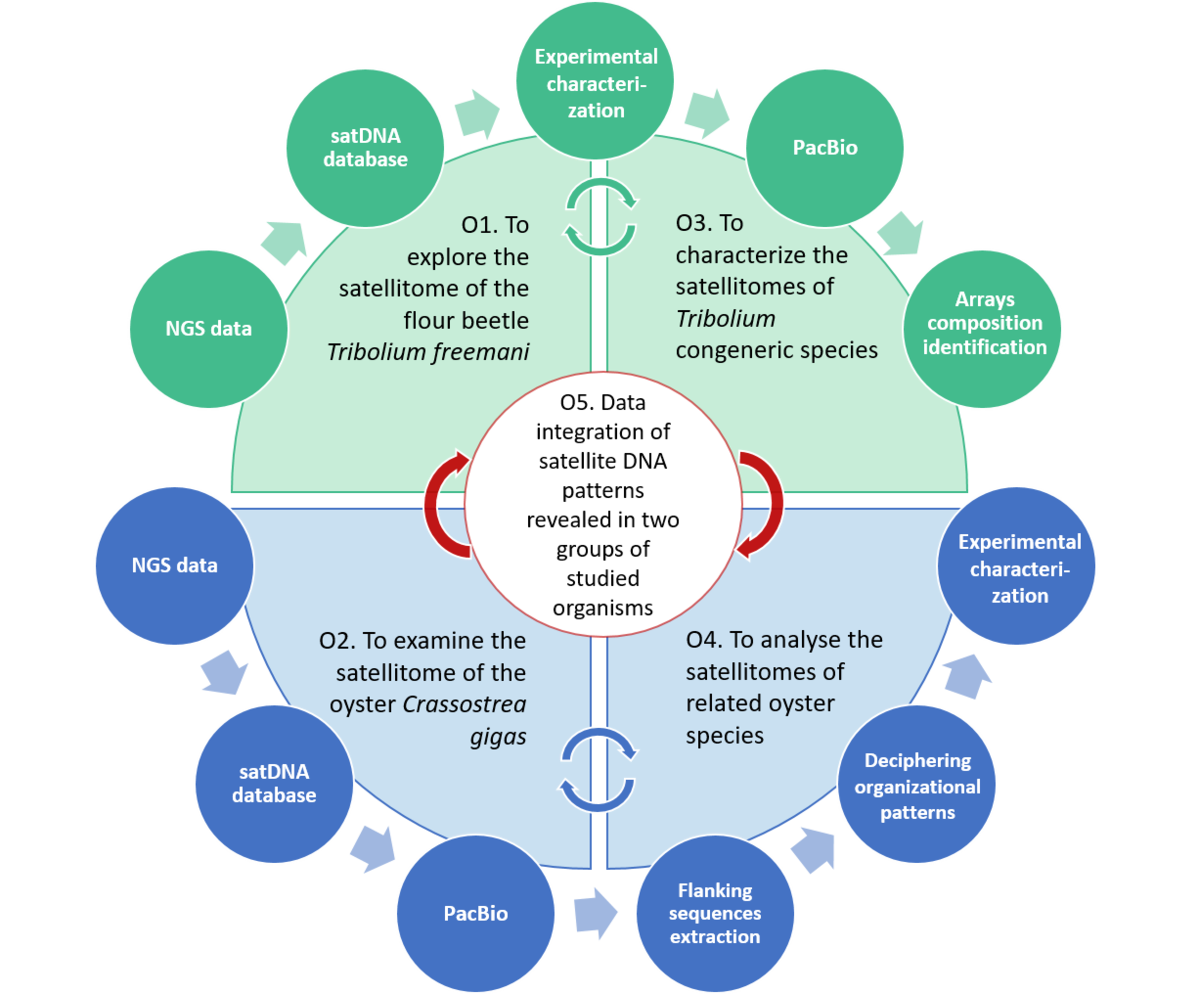
The main goal of this project is to investigate the satellitomes in two groups of invertebrates: (i) beetles of the genus Tribolium, characterized by highly abundant satDNAs located in large heterochromatic chromosomal domains, and (ii) bivalve species, known to have low satDNA contribution and chromosomes mostly devoid of large heterochromatic blocks. By engaging Illumina and PacBio sequencing combined with experimental methods to study satellitomes of eight species, our work addresses: (i) shared and species-specific satDNAs, (ii) satDNA sequence dynamics and chromosomal distribution in the analysed satellitomes, (iii) sequential patterns of satellite repeats, (iv) existence and conservation of potentially functional motifs in satDNA sequences, and (v) satDNA similarities and association with transposable elements (TEs).
By comprehensive surveys of satDNA concepts in two different invertebrate groups and ensuing data interlacing, we expect to further current ideas about satDNA structure and evolution, including the life-cycle, origin and destiny of satellite repeats, links between satDNAs and TEs, as well as possible mechanisms involved in the formation and spread of tandem repeats.
MODEL ORGANISMS
SCHEMATIC PRESENTATION OF THE PROJECT OBJECTIVES AND METHODOLOGICAL APPROACHES
PUBLICATIONS
Tunjić-Cvitanić M, García-Souto D, Pasantes JJ, Šatović-Vukšić E (2024) Dominance of transposable element-related satDNAs results in great complexity of "satDNA library" and invokes the extension towards "repetitive DNA library". Marine Life Science & Technology 6:236-251. doi.org/10.1007/s42995-024-00218-0
Volarić M, Despot-Slade E, Veseljak D, Pavlek M, Vojvoda Zeljko T, Mravinac B, Meštrović N (2024) The Genome Organization of 5S rRNA Genes in the Model Organism Tribolium castaneum and Its Sibling Species Tribolium freemani. Genes 15:776. doi.org/10.3390/genes15060776
Gržan T, Dombi M, Despot-Slade E, Veseljak D, Volarić M, Meštrović N, Plohl M, Mravinac B (2023) The Low-Copy-Number Satellite DNAs of the Model Beetle Tribolium castaneum. Genes 14:999. doi.org/10.3390/genes14050999
Šatović-Vukšić E, Plohl M (2023) Satellite DNAs-From Localized to Highly Dispersed Genome Components. Genes 14:742. doi.org/10.3390/genes14030742
Volarić M, Despot-Slade E, Veseljak D, Meštrović N, Mravinac B (2022) Reference-guided de novo genome assembly of the flour beetle Tribolium freemani. Int. J. Mol. Sci. 23:5869. doi.org/10.3390/ijms23115869
Šatović-Vukšić E, Plohl M. (2021) Classification problems of repetitive DNA Sequences. DNA 1:84–90. doi.org/10.3390/dna1020009
Volarić M, Veseljak D, Mravinac B, Meštrović N, Despot-Slade E (2021) Isolation of high molecular weight DNA from the model beetle Tribolium for Nanopore sequencing. Genes 12:1114. doi.org/10.3390/genes12081114
Tunjić-Cvitanić M, Pasantes JJ, García-Souto D, Cvitanić T, Plohl M, Šatović-Vukšić E. (2021) Satellitome analysis of the pacific oyster Crassostrea gigas reveals new pattern of satellite DNA organization, highly scattered across the genome. Int J Mol Sci. 22:6798. doi.org/10.3390/ijms22136798.
Team members
- Brankica Mravinac, PhD, Principal Investigator, IRB
- Miroslav Plohl, PhD, IRB
- Eva Šatović, PhD, IRB
- Tanja Vojvoda Zeljko, PhD, IRB
- Monika Tunjić Cvitanić, IRB
- Damira Veseljak, IRB
- Juan J. Pasantes, PhD, University of Vigo
- Daniel García Souto, PhD, University of Vigo