The ubiquitous and at the same time the least understood components of every eukaryotic genome are repetitive DNA sequences. They are divided into two groups: satellite DNAs (satDNAs), composed of sequences repeated in tandem, and transposable elements (TEs), interspersed throughout the genome. SatDNAs and TEs are considered to be crucial builders of genome architecture and drivers of its evolution, taking into consideration that genome evolution is strongly impacted by processes that reorganize repetitive DNA sequences and change their copy number. SatDNAs and their transcripts have active roles in heterochromatin establishment and centromere function, as well as in gene expression regulation under various biological contexts, e.g. stress response and environmental adaptation. TEs account for the majority of cis-regulatory DNA in the human genome, and have yielded numerous proteins co-opted for mammalian physiology and development. TEs extensively remodel regulatory networks that are involved in specific processes including dosage compensation, immunity, and early embryonic development, and can influence speciation. Their movement, regulatory activities, and effects on genome integrity also cause and exacerbate the effects of many diseases. The “mobilome” contributes strongly to eukaryote genome plasticity through bursts of activity. An increasing number of reports also show that satDNAs and TEs are tightly connected in many different ways.
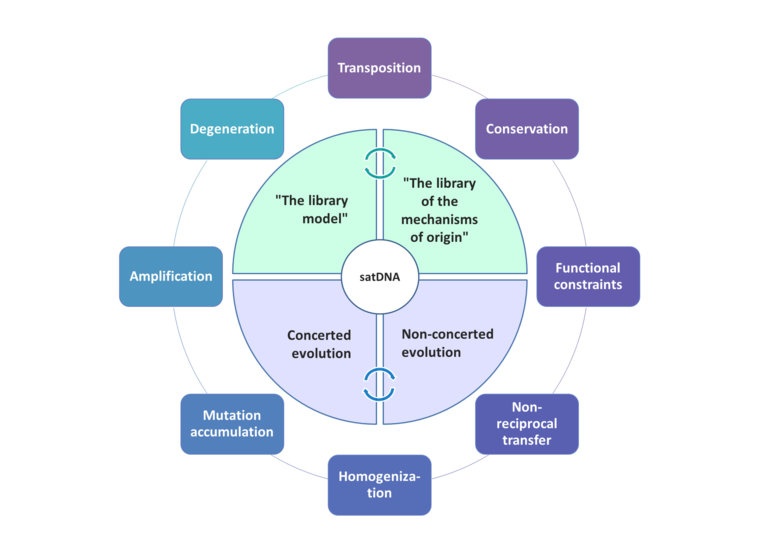
Evolution of satDNA sequences is influenced by many molecular mechanisms which underlie their sequence complexity, homogeneity, array size variability, and rapid evolution. High sequence homogeneity within a family of satDNA monomers is a consequence of their non-independent evolution, a phenomenon called “concerted evolution”. It is achieved through a process in which mutations are homogenized (either spread or eliminated) among members of a satDNA family in a genome, and concomitantly fixed within a group of reproductively linked organisms. Therefore, level of sequence variability in a satDNA family is an equilibrium between the accumulation of mutations, and the rate of their spread/elimination among satDNA monomers. In continuation, satDNA evolution follows the “library model” which proposes that closely related species share a set of satDNA families that originated from the common ancestor, with every satDNA being differentially amplified in each of the species. Thus, depending on the sequence dynamics in a particular organism or group of organisms, different evolutionary scenarios may occur, ultimately defining the overall satDNA landscape.
Schematic presentation of different mechanisms and major principles governing/influencing satDNA evolution.
(click for larger image)
Bivalve mollusks hold great economic and ecological importance. Their commercial significance is unquestionable in aquaculture, where they have several million-ton productions per year. The ecological impact of these organisms is emphasized when invasive bivalve species start to occupy new environments, significantly affecting native organisms in the new habitat. The research interest encompassing all aspects of bivalve biology is fast-growing, and their qualities are employed in many research areas, e.g. in stem cells differentiation, the ability to fight pathogens in the absence of adaptive immunity, as a source of alternative drugs, in mucosal immunity, toxicology, and even in cancer resistance. Recent studies have been moving towards genome-wide analyses (transcriptome, methylome, proteome, metabolome, epigenome, repeatome, satellitome, etc).
Repetitive sequences of oyster species have shown new patterns and exceptions from the canonical concepts, providing valuable contributions to the repetitive DNA biology. In addition, satDNAs in these organisms were shown to be intensively linked to TEs, especially with Helitron elements.
Employing different experimental and bioinformatic approaches, this research intends to explore the role of TEs in organization, propagation and evolution of satDNAs across a set of oyster species. We will examine the extent of “concerted evolution” and compare it between standalone and TE-incorporated satDNA arrays, both within and between species. The difference in the heterochromatin amount and content will be explored, and active parts of oyster mobilomes identified and compared. The study's findings will significantly advance our knowledge of repetitive genomic landscapes and their influence on chromosome structure. Importantly, comparative repeatomics will provide insights into the evolutionary processes and relationships of repetitive sequences in related species.
Team members:
dr. sc. Eva Šatović Vukšić
dr. sc. Miroslav Plohl
mag. educ. biol. et chem. Patrik Majcen
dr. sc. Juan J. Pasantes
dr. sc. Daniel García Souto
dr. sc. Andrea Luchetti
Contact:
Eva Šatović Vukšić esatovic@irb.hr