The team, led by Dr. Ivica Kopriva from the Laboratory for Machine Learning and Knowledge Representation, RBI Division of Electronics, in collaboration with Photon etc. (Canada), pathologists from University Hospital Dubrava and University Hospital Centre Zagreb, colleagues from the RBI Division of Molecular Medicine, and the Technical University of Munich, Germany, developed a computational method for analyzing hyperspectral images of histopathological samples. This method enables precise delineation of colorectal tumor tissue metastasized in the liver. The system distinguishes cancerous from healthy cells at the pixel level with more than 96% accuracy. Unlike standard AI methods, this algorithm requires only about 1% of manually labeled data from pathologists.
“Unlike conventional machine learning and deep learning methods, the segmentation algorithm we developed, based on Grassmann manifolds, works in a semi-supervised mode and requires only 1% of pathologist-labeled data,” explains Dr. Ivica Kopriva.
Hyperspectral Imaging of Histopathological Samples
Computational pathology methods have traditionally relied on RGB (color) images of histopathological samples, meaning they use only information visible to the human eye. This results in a loss of valuable data beyond the visible spectrum. Therefore, hyperspectral imaging—technology that reveals a much broader range of information and opens new possibilities in medicine—is increasingly being applied.
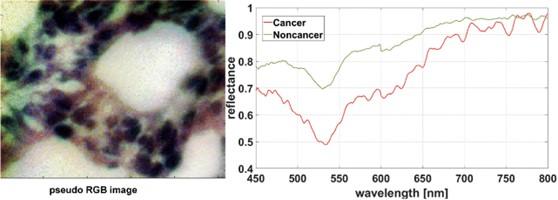
Left: pseudo-RGB image of a histopathological sample. Right: Typical spectral profile of tumor (red) and non-tumor tissue (green). Spectra averaged over a 5×5 pixel block.
Computational Method for Semantic Segmentation of Hyperspectral Images
The preparation process of histopathological samples can cause variations in image quality. For conventional color images, algorithms exist to correct such differences, but no such tools are available yet for hyperspectral images. As a result, the same tissue types can appear differently, even within the same image, complicating computational analysis. This also makes training AI models to recognize and separate tumor from healthy cells much more difficult. Moreover, training deep networks requires large datasets labeled at the pixel level by multiple pathologists. This is a major barrier to applying such methods for precise tissue classification.
To overcome these challenges, the team developed a new AI method capable of learning from a very small number of labeled examples. The method uses both colors (spectra) and shapes in tissue, accurately distinguishing tumor from healthy cells even within a single image. Pathologists need to label only about 1% of the pixels, after which the algorithm autonomously classifies the rest, reliably identifying tumor and non-tumor regions. This greatly simplifies training and reduces the need for extensive manual labeling—one of the main obstacles in applying deep learning.
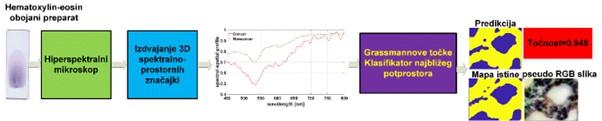
This method is easier to train, achieves performance comparable to advanced deep networks, and can already be applied at the level of individual images. This paves the way for faster and more practical integration into clinical settings. The developed methodology is schematically shown in Figure 3.
Illustration of the Grassmann manifold method in semi-supervised classification of hyperspectral images of frozen sections of colorectal liver metastases. Yellow indicates cancerous tissue, blue indicates non-cancerous tissue.
“To test the method, over eight years our team collected liver samples from 19 patients with colorectal cancer metastases during surgeries at University Hospital Dubrava, Zagreb. Twenty-seven hyperspectral images of histopathological samples from 14 patients were recorded at Photon etc. in Montreal, Canada. Using pseudo-RGB images, pathologists labeled tumor and non-tumor pixels with specialized software. The developed model proved capable of identifying tumor regions even under highly challenging conditions, when imaging conditions varied from case to case,” explains Dr. Kopriva.
A Method That Analyzes Tissue Almost Pixel by Pixel
The results obtained by the scientists are highly convincing.
“The new method, called GM-TSSA, successfully detected tumor regions with over 96% accuracy, using only one percent of manually labeled pixels (0.5% per class). In other words, it significantly outperformed six different deep neural network architectures, which require far more labeled data. This makes our method potentially suitable for real clinical conditions, where there is often neither the time nor resources to label thousands of samples or train highly demanding models,” notes Dr. Kopriva.
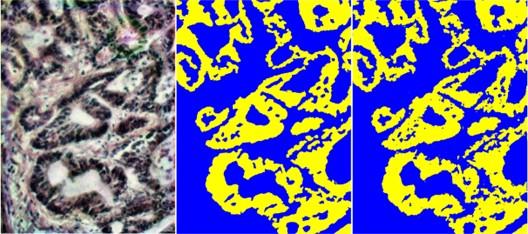
From left to right: pseudo-RGB histopathological image stained with hematoxylin and eosin, ground truth map derived from pixels labeled by five pathologists, semantic segmentation of a hyperspectral image using the Grassmann manifold method. Yellow indicates cancerous tissue, blue indicates non-cancerous tissue.
A key strength of this project lies in its openness. The entire dataset has been published in the RBI repository, while the source code is available on GitHub. This allows other researchers to test, refine, or apply the method to other cancer types, accelerating further development and clinical translation. It also contributes to the training of so-called vision-language foundation models in computational pathology, a particularly challenging task due to the scarcity of pixel-level annotated hyperspectral datasets.
Ultimately, hyperspectral imaging combined with image analysis algorithms has the potential to assist surgeons during operations. This could mean more precise assessment of resection margins and longer post-surgery survival for patients.
A Collaborative Effort with International Impact
The research team brings together experts from multiple disciplines: computer science, physics, applied mathematics, medicine, and clinical practice. In addition to project leader Dr. Ivica Kopriva (RBI), important contributions came from:
- Dr. Dario Sitnik (Technical University of Munich);
- Dr. Laura-Isabelle Dion-Bertrand, formerly at Photon etc. Canada (now at the Department of Applied Physics, University of Montreal);
- Pathologists Dr. Arijana Pačić and the late Dr. Gorana Aralica (University Hospital Dubrava, Zagreb);
- Pathologist Dr. Marija Milković Periša (University Hospital Centre Zagreb and University of Zagreb School of Medicine);
- Dr. Mirko Hadžija and Dr. Marijana Popović Hadžija (RBI Division of Molecular Medicine).
International Recognition
The research has been published in the renowned scientific journal Computers in Biology and Medicine, ranked among the top 7% of journals in biology and the top 15% in computer science. Publishing in such a journal confirms the high scientific value and application potential of this method.
The project was funded by the Croatian Science Foundation under grant HRZZ-IP-2022-10-6403.